6| Rules of Inheritance
Basic Mendelian Genetics
Although a background in genetics is certainly not essential for appreciation of many populational and ecological phenomena, it is a useful aid for application to some such phenomena and is required for a full understanding of others. Precise rules of inheritance were actually unknown when Darwin (1859) developed the theory of natural selection, but they were formulated a short time afterward (Mendel 1865). Darwin accepted the hypothesis of inheritance in vogue at the time: blending inheritance. Under the blending inheritance hypothesis, genetic makeups of both parents are imagined to be blended in their progeny, and all offspring produced by sexual reproduction should be genetically intermediate between their parents; genetic variability is thus lost rapidly unless new variation is continually being produced. (Under blending inheritance and random mating, genetic variability is halved each generation.) Darwin was forced to postulate extremely high mutation rates to maintain the genetic variability observed in most organisms, and he was painfully aware of the inadequacy of knowledge on inheritance. Mendel's discovery of particulate inheritance represents one of the major empirical breakthroughs in biology.
Mendel performed breeding experiments with different varieties of peas, paying close attention to a single trait at a time. He had two types that bred "true" for yellow and green peas, respectively. When a purebred green pea plant was crossed with a purebred yellow pea plant, all progeny, or individuals of the first filial generation (F1), had yellow peas. However, when these F1 plants were crossed with each other or self-fertilized, about one out of every four offspring in the second filial generation (F2) had green peas. Furthermore, only about one-third of the yellow F2 pea plants bred true; the other two-thirds, when self-fertilized, produced some offspring with green peas. All green pea plants bred true. Mendel proposed a very simple quantitative hypothesis to explain his results and performed many other breeding experiments on a variety of other traits that corroborated and confirmed his interpretations. Subsequent work has strengthened Mendel's hypothesis, although it has also led to certain modifications and improvements.

Modern terminology for various aspects of Mendelian inheritance is as follows: (1) the “character” or “dose” controlling a particular trait is termed an allele; (2) its position on a chromosome (defined later) is termed its locus; (3) a single dose is the haploid condition, designated by n, whereas the double-dosed condition, designated by 2n, is diploid (polyploids, such as triploids and tetraploids, are designated with still higher numbers); (4) the set of alternative alleles that may occur at a given locus (there can be only two alleles in an individual, but there may be more than two in any given population) is termed
a gene; (5) purebred diploid individuals with identical alleles are homozygotes, homozygous for the trait concerned; (6) individuals with two different alleles, such as the preceding F2 plants, are heterozygotes, heterozygous at that locus; (7) an allele that masks the expression of another allele is said to be dominant, whereas the one that is masked is recessive; (8) unlinked alleles separate, or segregate, from each other in the formation of gametes; (9) whenever heterozygotes or two individuals that are homozygous for different alleles mate, new combinations of alleles arise in the following generation by reassortment of the genetic material; (10) observable traits of an individual (e.g., yellow or green in the previous example) are aspects of its phenotype, which includes all observable characteristics of an organism; and (11) whether or not an organism breeds true is determined by its genotype, which is the sum total of all its genes.
Occasionally, some organisms have pairs of alleles with incomplete dominance. In such cases, the phenotype of the heterozygote is intermediate between that of the two homozygotes; that is, phenotype accurately reflects genotype and vice versa. Presumably, alleles conferring advantages upon their bearers usually evolve dominance over time because such dominance ensures that a maximal number of the organism's progeny and descendants will benefit from possession of that allele. The apparent rarity of incomplete dominance is further evidence that dominance has evolved. So-called wild-type alleles, that is, those most prevalent in natural wild populations, are nearly always dominant over other alleles occurring at the same locus. Geneticists have developed numerous theories of the evolution of dominance, but the exact details of the process have not yet been completely resolved.
Mendel postulated that each pea plant had a double dose of the "character" controlling pea color but that only a single dose was transmitted into each of its sexual cells, or gametes (pollen and ovules or sperm and eggs). Purebred plants, with identical doses, produced genetically identical single-dosed gametes; the above-mentioned
F1 plants, on the other hand, with two different doses, produced equal numbers of the two kinds of gametes, half bearing the character for green and half that for yellow. In addition, Mendel proposed that yellow masked green whenever the two occurred together in double dose; hence all F1 plants had yellow peas, but when self-fertilized, produced some F2 progeny with green peas. All green pea plants, which had a double dose of green, always bred true.
Cytological observations of appropriately prepared cell nuclei confirm Mendel's hypothesis beautifully (Figure 6.1). Microscopic examination of such cells reveals elongated dense bodies in cell nuclei; these are chromosomes, which contain actual genetic material, DNA. Nuclei of diploid cells, including the zygote (the fertilized ovule or egg) and the somatic (body) cells of most organisms, always contain an even number of chromosomes. (The exact number varies widely from species to species, with as few as two in certain arthropods to hundreds in some plants.) Pairs of distinctly similar homologous chromosomes are always present and often easily detected visually. However, gametes contain only half the number of chromosomes found in diploid cells, and, except in polyploids, none of them are homologous. Thus, haploid cells contain only one full set of different chromosomes and alleles, or one genome, whereas all diploid cells contain two. During the reduction division (meiosis) in which diploid gonadal cells give rise to haploid gametes, homologous chromosomes separate (Figure 6.1). Later, when male and female gametes fuse to form a diploid zygote that will develop into a new diploid organism, homologous chromosomes come together again. Hence, one genome in every diploid organism is of paternal origin while the other is of maternal ancestry. Because each member of a pair of homologous chromosomes separates from its homologue independently of other chromosome pairs, the previous generation's chromosomes are reassorted with each reduction division. Thus, the genetic material is regularly rearranged and mixed up by the dual processes of meiosis and the actual fusion of gametes (this has been termed the "Mendelian lottery").
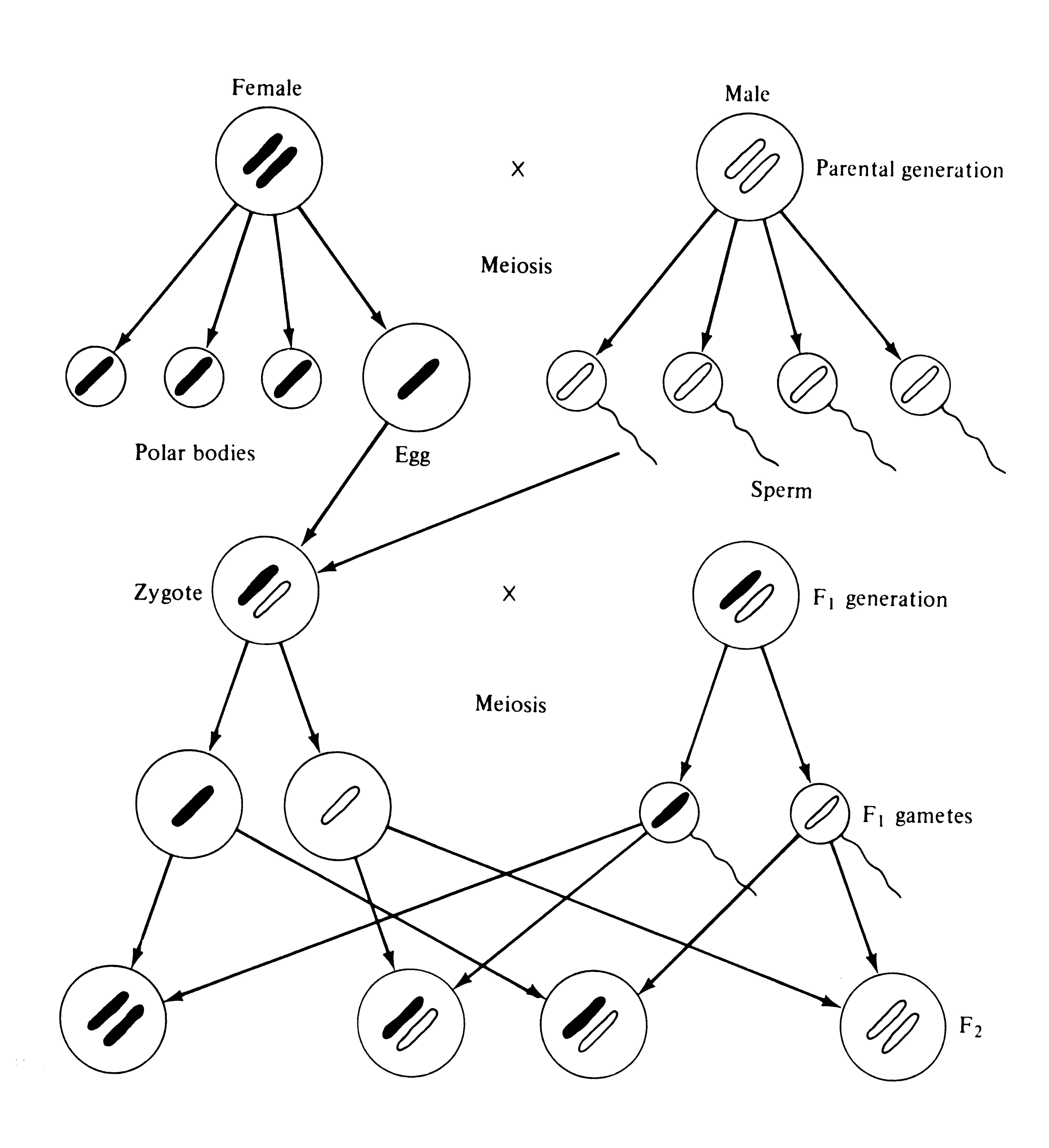
-
Figure 6.1. Diagrammatic representation of the cytological events in cell nuclei showing how the two parental genomes are sorted and recombined in the next generation, or the F2. For simplicity, only one pair of chromosomes is shown and the complex events of the reduction division (meiosis) are omitted.
Numerous different loci and allelic systems occur on each chromosome. Two different traits controlled by different alleles located on the same chromosome do not segregate truly independently but are statistically associated with or dissociated from one another. This is the phenomenon of linkage. During meiosis, homologous chromosomes can effectively exchange portions by means of crossovers; this process is referred to as recombination. Because the frequency of occurrence of crossovers between two loci increases with the distance between them on the chromosome,
geneticists use crossover frequencies to map the effective distance between loci, as well as their positions relative to one another on chromosomes. By means of close linkage, whole blocks of statistically associated alleles can be passed on to progeny as a functionally integrated unit of coadapted alleles.
Certain kinds of chromosomal rearrangements, such as inversions, may suppress crossovers. Indeed, a major advantage of chromosomes is that they enhance the degree to which clusters of genes can occur together. In many organisms a single pair of chromosomes, termed sex chromosomes, determine the sex of their bearer (the remaining chromosomes, which are not involved in sex determination, are autosomes). Typically one homologue of the sex chromosome pair is smaller. In the diploid state, an individual heterozygous for the sex chromosomes is heterogametic. In mammals, males are the heterogametic sex with an XY pair of sex chromosomes, whereas females are the homogametic sex with an XX pair. Because male-male matings are impossible, the homozygous genotype YY can never occur. In birds and some other organisms, the female is the heterogametic sex. In many reptiles, sex chromosomes do not exist and sex is determined by the environmental temperature at which eggs are incubated.
Although natural selection actually operates on phenotypes of individuals
(i.e., an organism's immediate fitness is determined by its total phenotype), the effectiveness of selection in changing the composition of a population depends on the heritability of phenotypic characteristics or the percentage of phenotypic variability attributable to genotype.
Traits that are under strong selection usually display low heritability because the genetic component of phenotypic variability has been reduced by selection. Because nongenetic traits are not inherited, differential reproduction by different phenotypes stemming from such nontransmittable traits obviously cannot alter a population's gene pool. Different genotypes may often have fairly similar phenotypes and thus similar fitnesses. Selection may even favor alleles that are "good mixers" and work well with a wide variety of other genes to increase their bearer's fitness in various genetic backgrounds (Mayr 1959). Conversely, of course, identical genotypes can develop into rather different phenotypes under different environmental conditions.
Genes that act to control the expression of other genes at different loci are called modifier or regulatory genes, whereas those that code for specific cell products are termed structural genes (some genes do not fit this dichotomy but may serve in both capacities). Although relatively little is known, geneticists imagine that an intricate hierarchy must exist leading from regulator genes to structural genes to proteins to other non-proteinaceous metabolic products to specific phenotypes. Moreover, complex interactions must occur among regulators as they do among proteins and other metabolites such as neurotransmitters and hormones.
Humans and the great apes share many genes, including the familiar ABO blood groups. A detailed molecular comparison of human proteins with those of chimpanzees by three different techniques (sequencing, immunological distance, and electrophoresis) revealed nearly identical amino acid sequences in the vast majority of proteins (99 percent similarity, with concordant results at 44 loci), presumably the products of structural genes (King and Wilson 1975). Apparently, a relatively few genetic changes in major regulatory genes can have profound phenotypic effects without much difference at the level of proteins. Thus, relatively trivial genetic differences can lead to major phenotypic differences. The genetic similarity between humans and chimps is comparable to that of sibling species in insects and other mammals. Recent DNA hybridization studies have demonstrated 98.4 percent similarity between humans and chimpanzees (Sibley et al. 1990). Whereas humans are placed in the family Hominidae, chimps are placed with the great apes in the family Pongidae. However, phylogenies based on molecular similarities show that humans are embedded within the apes (Figure 6.2). Clearly, it is time to consider reclassification!
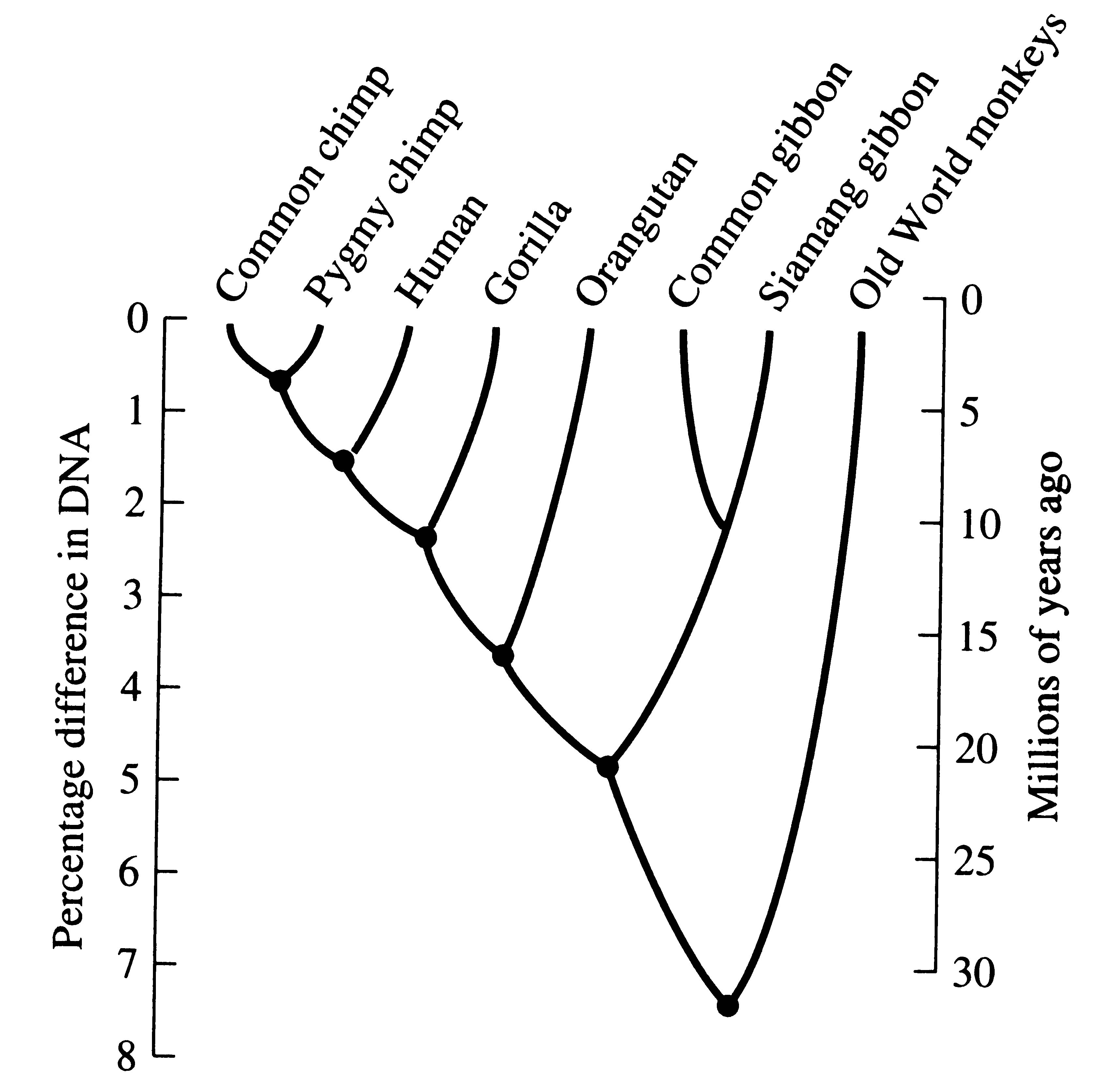
-
Figure 6.2. Phylogeny of primates based on DNA hybridization. [Adapted from Diamond (1991) after Sibley and Ahlquist.]
Nature versus Nurture
A widespread misconception is that any phenotypic trait can always be assigned to either one of two mutually exclusive categories: genetic or environmental. However, this dichotomy is not only oversimplified but can be rather misleading. Because natural selection acts only on heritable phenotypic traits, even environmentally flexible traits must usually have an underlying genetic basis. For example, when grown on dry plant foods, the Texas grasshoppers Syrbula and Chortophaga become brown, but when fed on moist grasses, these same insects develop green phenotypes -- this classic "environmentally induced" polymorphism is presumably highly adaptive since it produces background color-matching cryptic green grasshoppers when environments are green but brown ones in brown environments (Otte and Williams 1972). The capacity for developmental plasticity itself has almost surely evolved in response to the unpredictable environment these grasshoppers must face. If enough were known, much environmentally determined phenotypic variation would presumably have a somewhat comparable basis in natural selection. Thus, truly nongenetic traits are unimportant and uninteresting simply because they cannot evolve and do not affect fitness. Indeed, for purposes of evolutionary ecology, virtually all traits can be considered as being subject to natural selection (those that are not cannot easily persist and have little or no evolutionary significance). The complete set of different phenotypes that can be produced by a given genotype across a range of environments is called its reaction norm.
Selfish Genes
Certain alleles do not obey the Mendelian lottery of meiosis and recombination: instead these "outlaw genes" obtain disproportionate representation in a carrier's gametes at the expense of alternate alleles on homologous chromosomes. An example of such a selfish gene is the "segregation distortion" allele in the fruit fly Drosophila. Males heterozygous for this sex-linked trait produce sperm of which some 95 percent carry the allele (rather than the expected 50 percent). This process is known as meiotic drive. Why don't more genes behave in this manner? Very probably the intense contest for representation in the gametes has itself ended in a stalemate, yielding the traditional Mendelian ratios (viewed in this way, segregation itself is clearly a product of natural selection).
Advocates of the "selfish gene" hypothesis (Dawkins 1982, 1989) argue somewhat as follows. Barring mutations, genes are perfect replicators, always making exact copies of themselves. However, phenotypes of individual organisms are transmitted to their offspring only imperfectly, at least in sexually reproducing species. Individuals can thus be viewed as mere "vehicles" for the genes that they carry.
Except for viruses, genes usually do not exist in isolation but must occur together in large clusters. A sort of "packaging problem" arises, with the number of copies left by any given allele depending upon the particular combination of other genes, or "genetic background," in which the allele concerned actually occurs. (Actually, of course, selection "sees" the phenotypic expression resulting from interactions among all alleles in a particular genetic constellation in a given environment.) Also, genes clearly cannot replicate themselves except by means of successful reproduction of the entire organism. Hence selection must usually favor genes that work together to enhance an individual's ability to perpetuate them, thus increasing its fitness. A "parliment of genes" acts to govern the phenotype. A mutant gene that prevented its bearer from reproducing would be short-lived indeed! Likewise, mutations that reduce the reproductive success of individuals will normally be disfavored by natural selection. Viruses are indeed selfish genes, as their interests need not coincide with those of their hosts.
Population Genetics
Each generation, sexually reproducing organisms mix their genetic materials. Such shared genetic material is called a gene pool, and all the organisms contributing to a gene pool are collectively termed a Mendelian population. Gene pools have continuity through time, even as individuals are added and removed by births and deaths. One of the most fundamental concepts of population genetics is the notion of gene frequency. An allele's frequency in the haploid gene pool, or its proportional representation, is traditionally represented by the symbol p or q. Changes in allele frequencies in a gene pool in time constitute evolution. An individual's ability to contribute its own genes to the gene pool represents that individual's fitness.
Equilibrium frequencies of various diploid genotypes that emerge in a given gene pool, given random mating and no evolution, can be calculated from the haploidgene frequencies using the binomial expansion (also known as the Hardy-Weinberg equilibrium):
 1 = (p + q)(p + q) = p2 + 2pq + q2 1 = (p + q)(p + q) = p2 + 2pq + q2
If p is the frequency of allele A1 and q the frequency of allele A2, the expected equilibrium frequencies of the three diploid genotypes A1/A1, A1/A2, and A2/A2 are given by the three terms at the right: p2 , 2pq, and q2, respectively.
With random mating and no evolution, allele frequencies remain unchanged from generation to generation. The equilibrium frequency of heterozygotes reaches its maximum of 50 percent when the two alleles are equally represented (p = q = 0.5;
Figure 6.3).
-
Figure 6.3.Frequencies of the three diploid genotypes for various gene frequencies in a two-allele system in Hardy-Weinberg equilibrium.
Population geneticists have relaxed these limiting assumptions and elaborated and extended these equations to model various phenomena such as random genetic
drift, gene flow, nonrandom mating, and frequency-dependent selection. Genotypes can also be assigned fixed relative fitnesses; when the heterozygote is fitter than either homozygote, both alleles are maintained indefinitely, even if one or both of the homozygotes is actually lethal! This widespread and important phenomenon is known as heterozygote advantage or heterosis. Such a reduction in fitness due to genetic segregation is termed genetic load. If one homozygote enjoys the highest fitness, it is favored by natural selection until that locus becomes "fixed" with an allele frequency of 1.0. Ultimately, maintenance of genetic variation depends on the precise rules coupling allele frequencies to genotype frequencies.
Maintenance of Variability
The fundamental source of variation between individuals is sexual reproduction; reassortment and recombination of genes in each generation ensures that new genotypes will arise regularly in any population with genetic variability. In most higher organisms, no two individuals are genetically identical (except identical twins and progeny produced asexually). Population biologists are interested in understanding factors that create and maintain genetic variability in natural populations. When a population is reduced to a very small size, it must go through a genetic bottleneck which can greatly reduce genetic variability. Numerous genetic mechanisms, including linkage and heterosis, produce genetic variability both within and between populations.
At the outset, we must distinguish phenotypic from genotypic variation. The phenotypic component of variability is the total observable variability; the genotypic component is that with a genetic basis. It is usually difficult to distinguish genetically induced variation from environmentally induced variation. However, by growing clones of genetically identical individuals (i.e., with the same genotype) under differing environmental conditions, biologists have been able to determine how much interindividual variation is due to the developmental plasticity of a particular genotype in different environments. Pedigree studies show that approximately half the phenotypic variation in height observed in human populations has a genetic basis and the remaining variation is environmentally induced. The proportion of phenotypic variation that has a genetic basis is known as heritability. Because natural selection can act only on heritable traits, many phenotypic variants may have little direct selective value. The degree of developmental flexibility of a given phenotypic trait strongly influences an organism's fitness; such a trait is said to be canalized when the same phenotypic character is produced in a wide range of genetic and environmental backgrounds. Presumably, some genes are rather strongly canalized, such as those that produce "wild-type" individuals, whereas others are less determinant, allowing individuals to adapt and regulate via developmental plasticity. Such environmentally induced phenotypic varieties are common in plants, but they are less common among animals, probably because mobile organisms can easily select an appropriate environment. Presumably, it is selectively advantageous for certain genetically induced traits to be under tight control, whereas others increase individual fitness by allowing some flexibility of response to differing environmental influences.
Genotypic and phenotypic variation between individuals, in itself, is probably seldom selected for directly. But it may often arise and be maintained in a number of more or less indirect ways. Especially important are changing environments; in a temporally varying environment, selective pressures vary from time to time and the phenotype of highest fitness is always changing. There is inevitably some lag in response to selection, and organisms adapted to tolerate a wide range of conditions are frequently at an advantage. (Heterozygotes may often be better able to perform under a wider range of conditions than homozygotes.) Indeed, in unpredictably changing environments, reproductive success may usually be maximized by the production of offspring with a broad spectrum of phenotypes (which may well be the major advantage of sexual reproduction).
Similar considerations apply to spatially varying environments because phenotypes best able to exploit various "patches" usually differ. On a broader geographic level, differences from one habitat to the next presumably often result in different selective milieus and therefore in different gene pools adapted to local conditions. Gene flow between and among such divergent populations can result in substantial amounts of genetic variability, even at a single spot.
Competition among members of a population for preferred resources may often confer a relative advantage on variant individuals that are better able to exploit marginal resources; thus, competition within a population can directly favor an increase in its variability. By virtue of such variation between individuals, the population exploits a broader spectrum of resources more effectively and has a larger populational "niche breadth"; the "between phenotype" component of niche breadth is great (Roughgarden 1972).
Because such increased phenotypic variability between individuals promotes a broader populational niche, this has been termed the "niche-variation hypothesis" (Soulé and Stewart 1970). Similarly, environments with low availability of resources usually require that individuals exploiting them make use of a wide variety of available resources; in this case, however, because each individual must possess a broad niche, variation between individuals is not great (i.e., the "between-phenotype" component of niche breadth is slight, whereas the "within-phenotype" component is great).
One further way in which variability can be advantageous involves coevolutionary interactions between individuals belonging to different species, especially interspecific competition and predation. Fisher (l958b) likened such interspecific interactions and coevolution to a giant evolutionary game in which moves alternate with countermoves. It may well be more difficult to evolve against an unpredictable and variable polymorphic species than against a better standardized and more predictable monomorphic species. A possible example may be foraging birds developing a "search image" for prey items commonly encountered, often bypassing other less abundant kinds of suitable prey.
Units of Selection
Classical Darwinian natural selection acts only on heritable phenotypic traits of individuals. As discussed earlier, selfish genes are also known to exist. Can selection operate on entire groups of individuals such as families, colonies, populations, species, communities, and ecosystems? To what extent is the individual a natural "unit" of selection? How are conflicts between suborganismal, organismal, and superorganismal levels of selection resolved? These questions are often discussed both by geneticists and by ecologists, but there is no clear consensus as to correct answers.
Many behavioral and ecological attributes can be interpreted as having evolved for the benefit of the group rather than the individual. As an example of such group selection, consider the assertion that "mockingbirds lay fewer eggs during a drought year because competition for limited food supplies would be detrimental to the species." Such statements have a fatal flaw: "Cheaters" that laid as many eggs as possible would reap a higher reproductive success than individuals that voluntarily decreased their clutch size for the "benefit of the species." The same phenomenon can be interpreted more plausibly in terms of classical Darwinian selection at the level of the individual. During droughts, parental birds cannot bring as many insects to their nest and therefore cannot feed and fledge as many chicks as they can when food supplies are more ample. Birds can actually leave more surviving offspring to breed in the next generation by laying fewer eggs. Most evolutionary biologists now dismiss the preceding sort of "naive" group selection as untenable.
In the last two decades, thinking about group selection has achieved considerably greater sophistication, although it remains speculative. Two distinct types of selection at the level of groups emerge from these mathematical arguments. For "extinction" group selection to oppose natural selection at the level of the individual, isolated selfish subgroups must go extinct faster than selfishness arises within altruistic subgroups and most newly founded isolates must be altruistic. "Graded" group selection requires that distinct subpopulations contribute differentially to reproduction in a bigger population at large. In essence, entire groups must possess differential rates of survivorship or reproduction (i.e., differential fitness).
A major consideration is the extent to which an individual's own best interests are in conflict with those of the group to which it belongs. Ultimately, the frequency of occurrence of socially advantageous behaviors depends largely on the precise form of the trade-offs between group benefit(s) versus individual cost(s). Any individual sacrificing its own reproductive success for the benefit of a group is obviously at a selective disadvantage (within that group) to any other individual not making such a sacrifice. Classical Darwinian selection will always favor individuals that maximize their own reproductive success. Clearly, the course of selection acting within groups cannot be altered by selection operating between groups. Group selection requires very restricted conditions.
Williams (1966a) reemphasized, restated, and expanded the argument against naive group selection, pointing out that classical Darwinian selection at the level of the individual is adequate to explain most putatively "group-selected" attributes of populations and species, such as those suggested by Wynne-Edwards (1962) and Dunbar (1960, 1968, 1972). Williams reminds us that group selection has more conditions and is therefore a more onerous process than classical natural selection; furthermore, he urges that it be invoked only after the simpler explanation has clearly failed. Although group selection is certainly possible, it probably would not actually oppose natural selection at the individual level except under most unusual circumstances. A special form of selection at the level of the individual, kin selection, may frequently be the mechanism behind many phenomena interpreted as evidence for group selection. We will return to this issue from time to time in later chapters.
Genetic Engineering
Modern molecular biotechnological tools, such as restriction enzymes and gene splicing, now enable geneticists to transfer particular genes from one organism to another. For example, the firefly gene for luciferase has been successfully transferred to tobacco, resulting in transgenic bioluminescent plants. Human insulin and growth hormones are now routinely produced in chemostats of E. coli bacteria that have had human genes spliced into their genomes. Some researchers have even proposed using such transgenic bacteria as live vaccines (the genetically altered bacteria would live within humans and would confer resistance to particular diseases such as hepatitis). Such recombinant DNA technology has also enabled us to produce useful new life forms such as pollutant-eating bacteria that can help us to clean up what's left of our environment. Research is in progress to transfer nitrogen-fixing genes into crop plants.
There are legitimate concerns, however, about the safety of research on such man-made transgenic organisms, particularly the possibility of accidental release of virulent strains that might attack humans. Such concerns have been addressed by implementation of strict containment procedures for recombinant DNA products, as well as by selecting and creating host organisms for foreign DNA that are incapable of surviving outside the laboratory.
Obviously, genetically engineered organisms must eventually be designed for release into nature (indeed, genetically engineered tomatoes are now being grown commercially). Another concern is that genetically engineered organisms could have adverse effects on other species in natural ecosystems. We already have enough natural pests and certainly don't want to make any new ones!
Unfortunately, we still know far too little to engineer ecological systems intelligently (obviously genetic engineers should work hand in hand with ecological engineers). Still another problem is the human tendency to allow short-term financial returns to override long-term prospects.
Selected References
Basic Mendelian Genetics
Darlington and Mather (1949); Darwin (1859); Ehrlich and Holm (1963); Fisher (1930); Ford (1931, 1964); King and Wilson (1975); Maynard Smith (1958); Mayr (1959); Mendel (1865); Mettler and Gregg (1969); Sheppard (1959).
Nature versus Nurture
Bradshaw (1965); Clausen, Keck, and Hiesey (1948); Greene (1989); Otte and Williams (1972); Quinn (1987); Via et al. (1995)
Selfish Genes
Alexander and Borgia (1978); Dawkins (1982, 1989); Hamilton (1967); Leigh (1977); Orgel and Crick (1980).
Population Genetics
Crow (1986); Crow and Kimura (1970); Falconer (1981); Fisher (1930, 1958a); Ginzburg and Golenberg (1985); Haldane (1932, 1941, 1964); Hedrick (1983); Mayr (1959); Mettler and Gregg (1969); Wright (1931, 1968, 1969, 1977, 1978).
Maintenance of Variability
Ehrlich and Raven (1969); Fisher (l958b); Mettler and Gregg (1969); Roughgarden (1972); Somero (1969); Soulé and Stewart (1970); Van Valen (1965); Wilson and Bossert (1971).
Units of Selection
Alexander and Borgia (1978); Boorman and Levitt (1972, 1973); Brown (1966); Cole (1954b); Darlington (1971); Darnell (1970); Dawkins (1976, 1982); Dunbar (1960, 1968, 1972); Emerson (1960); Emlen (1973); Eshel (1972); Fisher (l958a); Gilpin (1975a); Leigh (1977); Levins (1970, 1975); Lewontin (1970); Maynard Smith (1964); Sober and Wilson (1998); Uyenoyama (1979); Van Valen (1971); Wade (1976, 1977, 1978); Wiens (1966); Williams (1966a, 1971); D. S. Wilson (1975, 1980, 1983); E. O. Wilson (1973, 1976); Wright (1931); Wynne-Edwards (1962, 1964, 1965a, b).
Genetic Engineering
Baba et al. (1992); Cockburn (1991); Tiedje et al (1989); Simonsen and Levin (1988).
|

